Background
The nuclear genome is inherited in sexually reproducing
species from both parents with recombination occurring during meiosis (Futumya
1986). Much of the nuclear DNA consists of non-protein coding regions and
heterochromatin. Protein coding regions vary from single copy loci to repetitive
arrays (gene families).Protein coding loci may be monitored for variation
using allozyme analysis (Murphy et al. 1990).
For molecular analysis it is useful to study specific
genomic regions. One such region is the ribosomal DNA (rDNA) gene family.
The rDNA gene family is a multigene family consisting of many copies (100-
500 in animals) of genes encoding for three ribosomal components 28S, 5.8S
and 18S (Black et al. 1989). In most eukaryotes, the 5' to 3' organization
of the gene family is an external transcribed spacer (ETS); the 18S gene;
an internally transcribed spacer (ITS1); the 5.8S gene; ITS2; 28S gene;
and the intergenic spacer (IGS) (Gerbi 1985). The IGS region is followed
by another copy of the gene family.
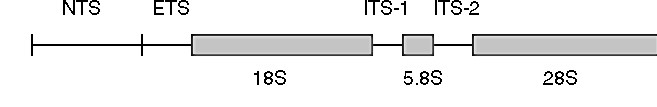
Figure 1. Diagram of the ribosomal DNA gene family
in animals (from Hillis & Davis 1986). The regions coding for the 5.8S,
18S, and 28S subunits of rRNA are shown by bars; NTS = non-transcribed
spacer, ETS = external transcribed spacer, ITS = internal transcribed spacer
regions.
Highly conserved regions in the ribosomal repeat
array can be used for study of relationships across phyla (Gerbi 1985),
more variable regions can be used at lower taxonomic levels. The ITS region
does not encode for any product, permitting it to evolve at a faster rate
than the ribosomal coding regions. The level of variation in this region
makes it suitable for detecting genetic variation among genera, species
and within species.
Size variation in the rDNA ITS region can be visualization
using gel electrophoresis
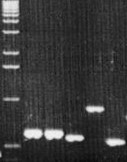
Figure
2. ITS1 size polymorphism in 11 tylenchid genera.
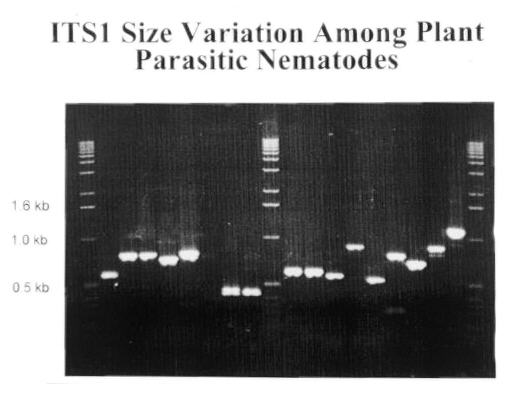 Figure 3. ITS1 size variation among Secernentean and
Adenophorean plant parasites. Here is a gel showing some the of size variation
that can occur. Amplicon is varying from 0.5 to 1.2 kb in size. This size
variation can be used as a quick screening technique for identifying plant
parasitic nematodes. Size variation within a genus may not be sufficient
for resolution on a agarose gel. To allow species identification Restriction
Fragment Length Polymorphisms (RFLP) are used.
Figure 3. ITS1 size variation among Secernentean and
Adenophorean plant parasites. Here is a gel showing some the of size variation
that can occur. Amplicon is varying from 0.5 to 1.2 kb in size. This size
variation can be used as a quick screening technique for identifying plant
parasitic nematodes. Size variation within a genus may not be sufficient
for resolution on a agarose gel. To allow species identification Restriction
Fragment Length Polymorphisms (RFLP) are used.
Technique
-
To measure the size of the amplicon, ensure that at molecular
size marker is run on the same gel as the amplicons. We like to use a 100
bp maker for our screening. Agarose gels are cast using 20 ml of 1% agarose
(We make up 200 ml of agarose solution at a time, using 2 g agarose, 200
ml 0.5% TBE and 6 ul of 10mg/ml ethidium bromide. This is made up in a
pyrex screw-top bottle and stored on the shelf. The solution is microwaved
for each gel casting).
-
Five ul of PCR product is combined with 0.5 ul loading buffer
and loaded on the gel.
-
The gel is placed in the gel box which contains a 0.5% TBE
solution and 6 ul of 10mg/ml ethidium bromide.
-
Gels are run for approximately 1 hour at 100 volts until
the bromphenol blue maker (migrates as fast as a 300 bp DNA fragment) has
reached the bottom of the gel.
-
Photograph the gel using instant b& w film. We use polaroid
film (ASA 3000) set at f8 aperture and 1 sec exposure.
-
We find the optimal way to measure amplicon size is to scan
the photo using a flatbed scanner into a computer then printing the image
at 300% larger using a laser printer.
Next, migration of the size marker fragments are measured
(usually the 1000 to 400 bp) in mm. The migration of the amplicons is measured
as well.
|